2015 DIMACS REU Proposed Project Descriptions
Please revisit this page frequently, additional projects will continue to be posted through January. An application can be updated by logging back into the account and including additional projects that are of interest.
Project #: DC2015-01
Mentor: Kim Hirshfield, MD, PhD, Rutgers Cancer Institute of New JerseyProject Title: Use of Comprehensive Genomic PRofiling for Personalized Cancer Therapy
Comprehensive genomic profiling of tumors through next-generation sequencing technologies offers the promise of personalized cancer therapy with targeted drugs. However, patient eligibility for genomically-guided drug therapies in the majority of clinical trials is linked to the presence of specific known or characterized genomic alterations. Through an ongoing clinical trial at Rutgers Cancer Institute of New Jersey, we have identified a number of patients whose tumors demonstrate genomic alterations in genes relevant to cancer biology or cancer therapy. While some patients have classic hot-spot mutations or well-characterized mutations, others display genomic findings with predicted altered function or with unknown consequences. We do not yet know how frequently uncharacterized alterations are found in comparison to characterized genomic alterations. Furthermore, for uncharacterized alterations that have been reported in cancer (e.g. in COSMIC or TCGA), we can then use on-line databases to help define which of the uncharacterized genomic alterations are most likely to have functionally-relevant consequences, e.g. through altered downstream gene expression or protein phosphorylation. This approach also offers a new avenue to identify novel mutations for which laboratory-based mutational characterization is most likely to yield data with clinical relevance. By establishing functionality of yet uncharacterized genomic alterations, the potential pool of patients who might benefit from targeted therapy could be increased.
References:Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502:333-9. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, et al. Cancer genome landscapes. Science. 2013; 339(6127): 1546-58. Carter C, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, et al. Cancer-specific High-throughput Annotation of Somatic Mutations: computational prediction of driver missense mutations. Cancer Res 2009; 69: 6660-6667.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-02
Mentor: David Foran, PhD, Rutgers Cancer Institute of New JerseyProject Title: High-throughput Analysis of Large Patient Cohorts
A major concentration for the Foran laboratory has been the development of a family of high-throughput, imaging, data-mining and computational tools for classifying malignancies and stratifying biomarker expression signatures from disease onset throughout progression. This work has led to several large-scale, inter-institutional investigative projects in bioinformatics, medical imaging, high-performance computing, computer-assisted diagnostics and therapy planning. Our team is utilizes these resources to conduct high-throughput analysis of large patient cohorts, store and mine large data sets; and determine the relationship between image analysis derived tumor information, gene expression category, patient stratification and clinical outcomes.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-03
Mentor: Yana Bromberg, PhD, Bromberg Labs, Biochemistry Microbiology, Rutgers UniversityProject Title: Mapping the Evolution of Redox Abilities
Energy is an organism's most basic need. The ability to carry out redox reactions, i.e. electron transfer, must have been among the first functionalities acquired by early life forms. Redox enzymes often use metals and metal-containing ligands. The goal of our work is thus to uncover how metal-containing enzymes, responsible for these critical electron transfer reactions, have evolved. The evolution of redox abilities can be mapped through the analysis of structural similarity of metal-binding protein folds, i.e. the active components of redox enzymes. From the RSCB PDB, we will obtain structures of transition metal binding protein folds, defined as all amino acids within a sphere of 15A radius from the center of the metal ligand. All-to-all structural alignments of metallospheres in our set will be used to optimize a function, differentiating functionally similar folds that bind the same ligand from all others. We will then align every metallosphere in our set (using our function similarity metrics) to every protein in the PDB. By analyzing the constructed similarity network for all protein spheres we will assess distant relations between redox proteins: whether they have common ancestors or if they developed independently. We also hope to understand whether metal binding folds in non-redox proteins may have come from a redox-fold ancestor. This study will contribute heavily to our understanding of the minimal structural requirements of redox, as well as illuminate evolutionary patterns leading to the current state of redox biology.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-04
Mentor: Brian Nakamura, PhD, CCICADA, Rutgers UniversityProject Title: Research in Enumerative/Algebraic Combinatorics
Enumerative combinatorics is an area of combinatorics that deals with the number of ways certain patterns can be formed. Most students are familiar with two simple examples of enumerative combinatorics: counting combinations and counting permutations. Algebraic combinatorics employs methods of abstract algebra, such as group theory and representation theory, within combinatorial contexts, as well as applying combinatorial techniques to algebra. Dr. Nakamura is a leading researcher with the Command Control Interoperability Center for Advanced Data Analysis (CCICADA) who research interests include enumerative and algebraic combinatorics, both from a theoretical and applied perspective. Dr. Nakamura will work with a qualified REU student to identify a new problem in enumerative/algebraic combinatorics that will be of interest to both the student and the mentor. This project will appeal to those students interested and applying algorithmic and computational techniques in a research setting.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-05
Mentor: Ned Lattime, PhD, Rutgers Cancer Institute of New JerseyProject Title: Whole Transcriptome Sequencing and Analyzing the Immune Tumor Microenvironment
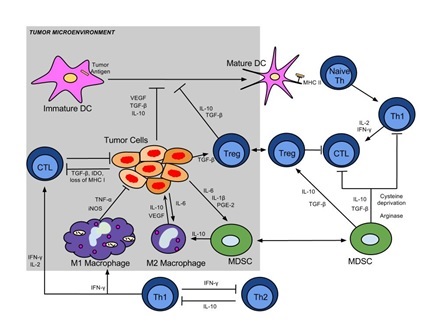
The overall goal of studies in our laboratory spanning preclinical animal models and clinical trials is the analysis of the tumor-immune interaction. Our studies involve the study of oncolytic viral vectors and vaccines and their effects in regulating host immunity with the ultimate goal of inducing an effective tumor specific immune response that will have activity against the primary tumor and metastastatic disease. As seen in the attached figure, the tumor-immune interaction is complex and invloves multiple immune cells and signalling pathways. We take advantage of whole transcriptome sequencing as a means of interrogating the noted pathways and evaluating the mechanistic results of our viral and vaccine interventions. The resultant large datasets require complex analyses and genomic expertise. We envision that students in the DIMACS program will have an opportunity to work with cell and immunology biologists and those with quantitative genomic expertise.
Project #: DC2015-06
Mentor: James Abello, PhD, abello at dimacs dot rutgers dot edu, DIMACSProject Title: Measuring the Mood of the Nation via Twitter - Project X
This project aims to create a data visualization and analytic platform to gauge public opinion on topics of current interests using messages on such social media venues as Twitter [1]. Students will explore a number of topics including public policy (deficit reduction, alternative energy, education), emergency response (police, firefighter), or critical services (health, evacuation) that grow out of social and economic needs of the students. Tweets matching a common topic will be collected and analyzed [2]. Keywords in the topic will be displayed on an interactive graph developed by students using geo-tags contained in the Tweets. Edge decomposition of the associated graph will be used to decompose the increasingly-complex graph over time into vertex groups of increasing minimum degree.
References:
[1] Turney P, "Thumbs Up or Thumbs Down? Semantic Orientation Applied to Unsupervised Classification of Reviews," Proceedings of the Association for Computational Linguistics (2002), pp. 417-424.
[2] Gansner E, Hu Y, and North S, "Visualizing Streaming Text Data with Dynamic Maps, to appear in Proceedings of the 20th International Symposium on Graph Drawing" (best paper award), (2012), TwitterScope website.
Project #: DC2015-07
Mentor: James Abello, PhD, abello at dimacs dot rutgers dot edu, DIMACSProject Title: Visualization of Time-Varying Graphs
ASK-GraphView is a node-link-based graph visualization system that allows clustering and interactive navigation of large graphs [1]. The system constructs a hierarchy on an arbitrary, weighted, undirected input graph using a series of increasingly sophisticated clustering algorithms. Building on this earlier work, this project will address the problem of visualizing graphs that are being collected in a streaming fashion. A key challenge is to identify graph sub-structures and statistics on them that can be used as the basis to depict evolution of the graph through time [2]. Another goal is to devise a visualization algorithm that produces a layout that is stable over time, thus preserving a user's mental map.
References:
[1] Abello J, Ham FV, Krishnan N, "AskGraphView: A Large Graph Visualization System", IEEE Transactions in Visualization and Computer Graphics, 12(5) (2006), 669-676.
[2] Hu Y, Kobourov S, and Veeramoni S, "Embedding, Clustering and Coloring for Dynamic Maps", Proceedings of IEEE Pacific Visualization Symposium, (2012).
Project #: DC2015-08
Mentor: James Abello, PhD, abello at dimacs dot rutgers dot edu, DIMACSProject Title: Detecting Anomolies in the Internet
This project will investigate algorithms for detecting anomalies in Internet traffic. Machine learning algorithms, in particular, implicit collaborative filtering [1], will be used to model normal Internet traffic records. Using this model, each machine is represented as a point in high dimension. Low-dimensional embedding algorithms [2] are then used to project points to 2 or 3D for visualization. Anomalies can be detected by visual inspection, as well as identified using clustering algorithms.
References:
[1] Hu YF, Koren Y, Volinsky C, "Collaborative Filtering for Implicit Feedback Datasets," IEEE International Conference on Data Mining (2008).
[2] Borg I and Groenen P, "Modern Multidimensional Scaling: theory and applications," New York: Springer-Verlag (1997).
Project #: DC2015-09
Mentor: Neil Campbell, PhD, Lattime Lab, Rutgers Cancer Institute of New JerseyProject Title: Micro-CT Optimization
The Preclinical Imaging Shared Resource at Rutgers Cancer Institute of New Jersey supports researchers as they study a wide variety of cancers and their potential treatment. One of the modalities that we use is micro-CT (computed tomography), which uses X-rays and computational processing to produce 3D images that can be manipulated in a range of ways. While a single micro-CT scan exposes an animal to an acceptable dose of ionizing radiation, longitudinal studies may, over repeated scans, raise that cumulative dose to clinically relevant levels. Using software, such as Matlab, and micro-CT raw data sets, we hope to identify combinations of acquisition settings and reconstruction algorithms that can allow us to reduce single scan time and exposure to ionizing radiation while providing optimal image quality.
Project #: DC2015-10
Mentor: Eric Allender, PhD, Computer Science, Rutgers UniversityProject Title: Boolean and Arithmetic Circuit Complexity
An n-ary function takes n inputs and produces some output. Some functions are easy to compute, whereas others will forever be difficult to compute, because they are inherently complex. Two useful measures of the complexity of a function are:
- Boolean circuit complexity (measuring the size of the smallest circuit of AND, OR and NOT gates that computes the function).
- Arithmetic circuit complexity (measuring the size of the smallest circuit of PLUS and TIMES gates that represents the function).
Project #: DC2015-11
Mentor: William Pottenger, PhD, DIMACS, Rutgers UniversityProject Title: Higher Order Learning
In our work we are developing a framework termed Higher Order Learning that builds models by leveraging associations in the form of latent relationships in an entity-relation graph. A general multi-attribute entity-relation graph has nodes that are people, places and things, etc. Higher Order Learning violates the underlying assumption made in traditional machine learning algorithms that records are independent and identically distributed(Taskar et al, 2002). Although this assumption simplifies the underlying mathematics of statistical models and of the corresponding parameter estimation procedures, it actually does not hold for many real world applications (Getoor & Diehl, 2005). Machine learning methods based on Higher Order Learning leverage relations between objects and features in an entity-relation graph and in doing so, operate on a much richer data representation compared to the traditional feature vector form. In this research, Higher Order Learning techniques have been investigated and applied to a number of interesting datasets including Border Gateway Protocol (cybersecurity), E-Commerce (web mining), and Nuclear Detection (homeland security). The approach has been shown to outperform the popular Support Vector Machine algorithm on benchmark textual data sets as well as real-world streaming data Ganiz, Lytkin & Pottenger). REU summer interns will have an unparalleled opportunity to explore Higher Order Learning algorithms and applications in the context of law enforcement and other domains.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-12
Mentor: William Pottenger, PhD, DIMACS, Rutgers UniversityProject Title: Entity Resolution
Emergencies invariably require crisis managers to field numerous calls for service such as 911, radio reports by police and fire teams, medical reporting, infrastructure support teams, etc. Technologies for Entity Resolution can be applied to handle the confusion by identifying geospatial, temporal, and content similarities in the messages, enabling first responders to resolve and identify the location and scope of multiple events. Entity Resolution also plays an important role in counter-terrorism, for example, in determining when two terrorist incidents are the same. This project will involve the research and development of Entity Resolution techniques to group incidents from the Internet Crime Complaint Center database (www.ic3.gov). REU summer interns will have an unparalleled opportunity to explore Entity Resolution algorithms and applications in the context of law enforcement and other domains.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-13
Mentor: Wilma Olson, Mary I. Bunting Professor, Chemistry & Chemical Biology, Rutgers UniversityProject Title: Effects of architectural and regulatory proteins on the spatial organization and expression of bacterial genes
Many genetic processes are controlled by proteins that bind at separate, often widely spaced, sites on DNA and hold the intervening double helix in a loop. Our group has initiated a series of computer simulations of the formation of the DNA loops implicated in the Escherichia coli lac operon, the classic textbook example of genetic action at a distance [1]. We incorporate effects of naturally occurring species found in the cell, such as the non-specific architectural protein HU [2] and the Lac repressor protein, on the looping free energies. By representing the DNA as a sequence of base pairs, we capture the structural details of protein-bound sites and the natural deformability of free DNA [3-5]. This approach allows for departures from conventional treatments of DNA, such as ideal elastic models, which may be divorced from reality, and leads to results that can be directly linked to genetic processing at the molecular level. We relate the computed looping propensities to DNA looping propensitieis derived from gene repression and single-molecule studies [6,7]. We are also collecting the simulated structures in a database of three-dimensional loop models that can be used as starting points in the determination of structures consistent with NMR and other local spectroscopic measurements and subsequently refined with these high-resolution data. The approach is general and can be applied to any known protein-mediated DNA looping systems as well as to the design of novel DNA constructs, such as the three-dimensional origami formed by looping DNA between carefully spaced four-way junctions [8].
References:
[1] Müller-Hill B (1996) The lac Operon. Walter de Gruyter, Berlin.
[2] Swinger KK, Lemberg KM, Zhang Y & Rice PA (2003) Flexible DNA bending in HU-DNA cocrystal structures. EMBO J 22, 3749-3760.
[3] Olson WK, Gorin AA, Lu X-J, Hock LM & Zhurkin VB (1998) DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc Natl Acad Sci USA 95, 11163-11168.
[4] Swigon D, Coleman, BD & Olson WK (2006) Modeling the Lac repressor-operator assembly: the influence of DNA looping on Lac repressor conformation. Proc Natl Acad Sci USA 103, 9879-9884.
[5] Czapla L, Swigon D & Olson WK (2008) Effects of the nucleoid protein HU on the structure, flexibility, and ring-closure properties of DNA deduced from Monte-Carlo simulations. J Mol Biol 382, 353-370.
[6] Becker NA, Kahn JD & Maher 3rd LJ (2005) Bacterial repression loops require enhanced DNA flexibility. J Mol Biol 349, 716-730.
[7] Han L, Garcia HG, Blumberg S, Towles KB, Beausang JF, et al.(2009) Concentration and length dependence of DNA looping in transcriptional regulation. PLoS ONE 4, e5621.
[8] Han D, Pal S, Nangreave J, Deng Z, Liu Y & Yan H (2011) DNA origami with complex curvatures in three-dimensional space. Science 332, 342-346.
Project #: DC2015-14
Mentor: Kevin Chen, PhD, Biology, Rutgers UniversityProject Title: Development of machine learning algorithms in biology
Hidden Markov Models (HMMs) are standard probabilistic models for time series or sequential data with many applications in computational biology, speech recognition and other applied fields. We are working to develop novel algorithms to learn the parameters of HMMs and related probabilistic models based on linear algebraic techniques which are faster and in some cases more accurate than traditional methods based on optimization techniques. We have developed a software tool implementing these methods and applied it to a data set of epigenomic marks across the human genome to understand the role of gene regulation in human diseases. This project has two components. The applied component is to write Python scripts to use the algorithm to understand the differences in gene regulation between different cell types (e.g. cancer cells vs. normal cells). The theoretical component is to extend the method to the more general "tree HMM" probabilistic model which allows the different cell types to be related by a tree. This project would be most suitable for a student with a strong background in advanced linear algebra, computer programming and probability/statistics.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-15
Mentor: Christopher Woodward (Math REU Coordinator), PhD, Mathematics, Rutgers UniversityProject Title: Projects in Pure Math
Several projects will be run in 2015 by members of the pure math faculty. Past topics have included Schubert calculus (Buch) Ricci flow(Song) mathematical physics and general relativity (Soffer) and mathematical Yang-Mills theory (Woodward).
Applicants interested in a project in pure mathematics should list this project in their application; further details will be worked out in the Spring.
Project #: DC2015-16
Mentor: Kostas Bekris, PhD, Computer Science, Rutgers UniversityProject Title: Algorithmic Challenges for Robots that Change their World
Robot manipulation is the capability of an autonomous system to alter its surroundings, such as grasping, pushing or throwing objects in its vicinity. Typically, it involves the use of robotic arms and hands but it can also be achieved by simpler mobile robots without complex end effectors. There are many issues that arise towards achieving efficient robot manipulation, which involve mapping the world, detecting objects and ways to interact with them. At the core of robot manipulation, however, lie very interesting representation and algorithmic challenges relating to task and motion planning. For instance, what is the state space of multiple arms transferring an object from an initial to a goal location? There are many locations where the arms can perform a handoff and many locations where the object can be potentially placed in an environment, which results in a combinatorial explosion of possible states that one can consider. Efficient state space representations are needed, given practical assumptions, in order to tackle such problems in an effective manner. Given compact state space representations, it is possible to consider appropriate search algorithms that look for the best sequence of operations that allow the solution of complex manipulation challenges. The student, who will work on this project, will work at an algorithmic and foundational level but also have the opportunity to simulate such challenges given existing open-source software tools, as well as perform experiments with a real robotic platform through interaction with graduate students in the research lab.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-17
Mentor: Christie Nelson, PhD, CCICADA, Rutgers UniversityProject Title: Modeling Social Media for Disaster Situations
Social media has many emerging outlets of communication that did not exist even fifteen years ago, including Twitter, Facebook, micro-blogs, etc. Social media and text messaging can be utilized for quick communication when time is of the essence, particularly during disasters. During a disaster type situation, could the way that social media and text messaging communication is currently done be improved? In particular, on the public side, is there a way to improve the wording of messages to aid in the automation of classification? Should a supervised or unsupervised approach be taken? A supervised approach uses a labeled training set to build a model, while an unsupervised approach does not. A project of joint interest to the mentor and to the student will be identified. Some background in programming is encouraged.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-18
Mentor: Jake Baron, CCICADA, Rutgers UniversityProject Title: Defining a Game Version of The Best Choice Problem
The best choice problem (also known as the secretary problem, the marriage problem, the sultan's dowery problem, the fussy suitor problem, or the googol game), has been studied extensively in the fields of applied probability, statistics, and decision theory. The basic form of the problem asks for an individual to make the best choice out of n rankable candidates for a position. The problem first appeared in print in a 1960 Martin Gardner Scientific American column in which he stated: "Ask someone to take as many slips of paper as he pleases, and on each slip write a different positive number. The numbers may range from small fractions of 1 to a number the size of a googol (1 followed by a hundred 0s) or even larger. These slips are turned face down and shuffled over the top of a table. One at a time you turn the slips face up. The aim is to stop turning when you come to the number that you guess to be the largest of the series. You cannot go back and pick a previously turned slip. If you turn over all the slips, then of course you must pick the last one turned." This project will concentrate on defining and solving a game version of this problem.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-19
Mentor: Jake Baron, CCICADA, Rutgers UniversityProject Title: The Numerical 3-Dimensional Matching problem in the case X=Y=Z
The Numerical 3-Dimensional Matching problem is a decision problem given by three multisets, X, Y, and Z, each containing , k elements and a bound b. The goal is to select a subset M of X×Y×Z such that every integer in X, Y, and Z occurs exactly once and that for every triple (x,y,z) in the subset x+y+z=b holds. Numerical 3-dimensional matching is an NP-complete decision problem. This project will explore the question: Does the Numerical 3-Dimensional Matching problem remain NP-hard in the case that X=Y=Z? This is a theoretical question that grew out of CCICADA's "BAM II" project for the US Coast Guard, about how to let Coast Guard stations share boats seasonally in order to reduce the overall number of boats needed.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-20
Mentor: Nina Fefferman, PhD, Ecology, Evolution, and Natural Resources and CCICADA, Rutgers UniversityProject Title: Developing and Applying Algorithms Inspired by Nature to Applications in Ecology and Epidemiology
Dr. Fefferman is a leading researcher with the Department of Ecology, Evolution, and Natural Resources and CCICADA who works largely in the areas of epidemiology, evolutionary & behavioral ecology, and conservation biology. Dr. Fefferman will work with a qualified REU student to identify a new problem from her area of research that will be of interest to both the student and the mentor. This project will appeal to those students who are interested in developing and applying mathematical and computational models to biological systems.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-21
Mentor: Eugene Fiorini, PhD, DIMACS, Rutgers UniversityProject Title: Research in Competition Graphs and Graph Forensics
Dr. Fiorini is a researcher with DIMACS who works largely in areas of graph theory. Dr. Fiorini will work with a qualified REU student to identify a new problem from his area of research that will be of interest to both the student and the mentor. This project will appeal to those students who are interested in competition graphs and graph theory applied to forensic science.
* You must be a U.S. Citizen or Permanent Resident to be eligible for this project.Project #: DC2015-22
Mentor: Christie Nelson, PhD, CCICADA, Rutgers UniversityProject Title: Experimental Designs for Walk Through Metal Detectors at Large Stadiums
When utilizing metal detectors at a large venue such as a sports stadium, there are the competing objectives of accuracy of the patron screening and the speed of throughput. This research, carried out in collaboration with large sports venues, analyzes the patron screening method of walk-through metal detectors ("walk-throughs").
Experimental design is the focus of the project, helping large venues better understand the performance of walk-throughs in real outdoor settings. Because of the number of experimental factors to be considered (type of item, location and height of object, orientation, speed of object passing through the machine, walk-through security setting, etc.), designing experiments require a sophisticated design tool called combinatorial experimental design. Experimental designs can focus on various questions, such as: does a walk-through catch each of the pre-specified prohibited test items; can multiple metal test objects can be hidden on a person and tested all at once in different height zones?
The student must be a US Citizen. In addition, statistics courses are preferred.
Project #: DC2015-23
Mentor: Gyan Bhanot, PhD, Rutgers Cancer Institute of New JerseyProject Title: Identifying Networks associated with Barretts Esophagus progression to Esophageal Cancer in the BAR-T cell model
Barretts esophagus is a pre-malignant condition found in people with GIRD (Gastro Intestinal Reflux Disease). Using RNA-seq data collected from immortalized Barretts cells treated with acid and bile for 70 weeks, the project involves identifying networks of genes which are disregulated as a function of time. The goal is to identify and study the gene networks in this model which are responsible for malignant transformation of Barretts cells into tumorigenic cells.